Myocardial fiber and connective tissue architectures are fundamental determinants of regional and global cardiac mechanical function. Structural alterations in the myocardial tissue matrix are apparent within 90 minutes of mild ischemic injury[1] and gross changes in the fiber and collagen network occur over a much longer time scale in infarcted tissue. A method for measuring intercellular structure might therefore be a sensitive indicator of the severity, extent and time course of ischemic injury, while also providing a means to study myocardial structure/function relationships.
Methods
MR Diffusion Imaging:
Isolated canine hearts were imaged using diffusion sensitive MRI and a
spin-echo acquisition sequence. Six sets of biphasic gradient pulses,
with diffusion sensitivity of B~300 s/mm^2, were used to
compute the diffusion tensor at each imaging voxel. The principal directions
of diffusion and their diffusion values were then calculated in
each voxel and the voxel locations registered in 3D.
An MR diffusion eigen-image was created by displaying the principal
direction corresponding to (and scaled by) the largest eigenvalue, as shown
in Figure 1.

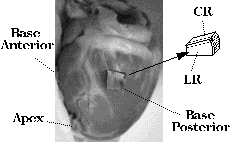
Histological analysis: Tissue blocks were harvested from 3 sites in the same heart used for the diffusion-sensitive imaging experiment. Transmural blocks of dimension ~1.5 cm^3 were excised, sectioned in orthogonal planes at 50--200micron thickness (see Figure 1) on a vibratome, transferred to a slide and viewed under low power (x10). Fiber angles were observed and recorded in sections cut orthogonal to the epicardial surface at ~1mm intervals.
| Apical Anterior | Basal Anterior |
 |
 |
| Basal Posterior | |
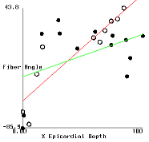 | |
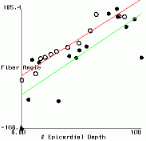
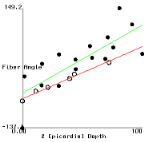
Figure 2 shows plots of fiber angle versus percent wall depth for the histological and MR-diffusion data. The nearly linear variation of fiber angle with depth correlates with previous findings[6]. Linear regression lines were fit to each data set at each depth. The fiber angle intercepts at the epicardium and endocardium and fitted slopes are presented in Table 1.
| (histo/MR) | apical ant. | basal ant. | basal post. |
| epi int. | -45.6/-89.1 | -66.8/-53.6 | -56.3/-30.3 |
|---|---|---|---|
| endo int. | 127/94 | 57/108 | 59/15 |
| slope | 173/183 | 124/162 | 115/46 |
Discussion
A correlation is observed between the computed direction of largest
in-plane diffusion and the histologically measured fiber angles in
at least 2 of the 3 regions analyzed.
The variance in the MR-based fiber angles is believed to arise from
the uncertainty
in the raw MR diffusion data, coupled with the relatively large MR voxel
size compared to the observed transmural fiber angle variations.
MR data from the extreme endo- and epi-cardial voxels are also likely
to exhibit corruption due to signal from the fluid surrounding the heart.
Fiber angle error at the intercepts is expected due to the uncertainty
in delimiting the endo- and epi-cardial borders from the MR images.
In the basal posterior site, where the slopes are least well correlated,
there is a correspondance between the histologically measured angles and
those from the MR analysis in the sub-epicardium where the distribution
of fiber angle was highly non-linear.
References
1. Zhao M et al,
2. Wong EC, Cox RW and Song AW
3. Hsu EW and Mori S,
4. Basser PJ, Mattiello J, and LeBihan D
5. Reese TG, Weisskoff RM, Smith RN, Rosen BR
6. Nielsen PMF, LeGrice IJ, Smaill BH, Hunter PJ
7. Legrice IJ, Smaill BH, Chai LZ, Edgar SG,
Acknowledgements
This research was funded through NIH HL41603, NSF BES-9634974 and the
Whittaker Foundation.